Cattle weight estimation model through readily photos
Sure! Here are **copy-paste-ready subtitles** for your blog website on **cattle weight estimation using photos**: ``` - Accurate Cattle Weight Estimation from Just a Photo - AI-Powered Livestock Monitoring Using Image Analysis - Estimating Cattle Weight Without Scales: Here's How - From Camera to Data: Smart Farming with Deep Learning - Image-Based Weight Prediction for Modern Cattle Farms - Revolutionizing Animal Weight Estimation with Computer Vision - Simple Photos, Smart Insights: Livestock Tech for the Future - Deep Learning Models for Cattle Weight Estimation Made Easy - Visual Data to Track Cattle Growth and Health - A New Era in Precision Farming Using AI and Photos ``` Let me know if you want more subtitle styles (e.g. formal, catchy, or keyword-rich for SEO).
Abstract
Cattle weight is a key indicator for assessing health, meat quality, productivity, and overall well-being. Manual measurement methods are time-consuming and induce stress reactions. In contrast, non-contact measurement methods based on computer vision can effectively and rapidly estimate cattle weight. These non-contact methods typically involve image processing and three-dimensional reconstruction, capturing image information with cameras and combining deep learning algorithms to estimate cattle weight. Despite their advantages, these methods are mainly designed for large-scale professional cattle farms and are characterized by high costs and complex operations, which do not meet the needs of individual cattle farmers. To address this issue, we propose the Lightweight Network-based Cattle Weight Estimation (LaWE) model, deployed on mobile phones and can measure cattle weight by capturing readily available photos. The LaWE model uses an optimized lightweight multi-scale fusion network to accurately extract keypoints from images and calculate relevant body measurements, which are then input into a specially designed deep regression network to estimate cattle weight. We conducted extensive experiments on both publicly available datasets and our self-collected dataset of 108 Horqin Yellow cattle. The results demonstrate that the LaWE model achieves an accuracy rate of over 97% in cattle weight estimation, meeting the requirements for low cost and real-time performance. Our research makes a significant contribution to the field of non-contact cattle weight estimation, providing practical technical support for smallholder farmers in the industry.
Measuring Tape

Introduction
By 2024, Chinese livestock husbandry accounted for 43.6% of the total output value of agriculture and animal husbandry, with cattle farming representing 17% of the national livestock husbandry. Based on this data, it can be inferred that the cattle breeding industry is a significant component of Chinese agriculture and animal husbandry. Therefore, ensuring the health, meat quality, and slaughter volume of cattle is a crucial task. Monitoring the condition of cattle during the breeding process is essential to achieve these goals. Cattle weight is an important indicator for assessing growth status, feeding efficiency, and health issues. Continuous weight gain indicates adequate nutrition intake and good health, while slow or stagnant weight gain may suggest insufficient feed supply or health problems, necessitating timely adjustments( Li et al. (2018), Smith et al. (2018)). Weight measurement also helps calculate feed efficiency, predict slaughter yield, and formulate optimal sales strategies, promoting the sustainable development of Chinese livestock husbandry.
Currently, cattle farming in China can be classified into two categories: professional breeding farms and individual cattle farmers, with individual cattle farmers accounting for 70% of the market share. Compared with professional farms, individual cattle farmers face difficulties in measuring the weight of cattle. Each cattle in a professional breeding farm is typically kept in a designated pen and has a specialized weighing area, making it convenient to lead the cattle into the weighing area and use modern technological equipment to measure their weight. For individual cattle farmers, professional weighing equipment is expensive, and individual farmers often adopt free-range model, with the cattle scattered over vast areas, lacking specific weighing areas. It is impractical to gather all the cattle in one place for centralized weighing, which requires a lot of manpower and time costs, and will bring pressure and fear to the cattle. Therefore, individual cattle farmers can only estimate the weight of cattle by visual inspection, which is not feasible for people without years of experience, and the error range of visual inspection is large. In such an environment, estimating the weight of cattle is a challenging task.

At present, scholars have conducted extensive research on non-ct methods for estimating livestock weight. The weight of cattle can be estimated by laser, pressure sensor and computer vision. Compared to manual measurement methods, these non-contact techniques improve efficiency and reduce the risks and errors associated with human intervention. Relevant studies have shown that computer vision methods can estimate weight through image or video processing and analysis. For example, Cominotte et al. (2023) have established multiple feeding troughs for cattle and installed data collection equipment above the feeding troughs. When the cattle is eating in the feed trough, take photos of the cattle back, use fully supervised segmentation and weakly supervised segmentation methods to statistically analyze features and estimate the weight of cattle. This method mainly involves weight analysis of the obtained images. Tu and Jørgensen (2023) proposed a visual analysis and prediction system that uses visual algorithms to determine areas related to pig weight, and then uses area to predict the live weight of newborn pigs. Cominotte et al. (2023) use depth cameras to obtain depth information of cattle, reconstructs three-dimensional images of cattle to obtain relevant biological features, and ultimately calculates the corresponding body weight. This method upgrades the technology for obtaining depth information, constructs a three-dimensional structure, and then estimates it to comprehensively and systematically obtain weight values.
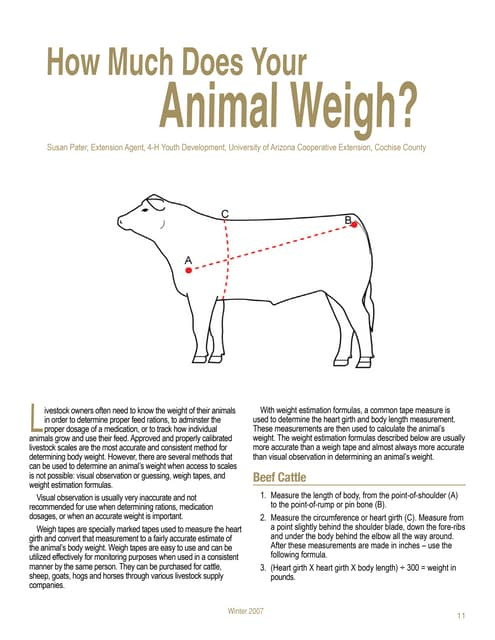
Based on the above research, it is known that using computer vision methods for livestock-related measurements has high accuracy and reliability, while reducing manual operations and time costs. However, the current methods are only applicable to fixed barn environments because they require the installation of measurement devices in specific areas and guiding livestock to those areas for measurements. In addition, the cost of professional equipment and measurement is usually relatively high. We conducted a survey on individual cattle farmers and found that the method of setting up fixed weighing station in specific areas is not feasible. The grazing mode of individual cattle farmers involves one person overseeing multiple cattle, and the fixed-area weighing method is too demanding for individuals in terms of workload. Individual cattle farmers graze their cattle in vast open areas and regularly change grazing zones. The fixed-area weighing method requires a stable network, but the data transmission services available in the grazing area are not sufficiently reliable. The equipment used for the fixed-area weighing method is expensive, and individual cattle farmers have limited financial resources.
Based on the aforementioned conditions, we propose a Lightweight Network-based Cattle Weight Estimation (LaWE) model. Firstly, we take into account the need for conveniently portable weight estimation devices for herders. We have found that mobile phones meet the requirements and can deploy the weight estimation model on them. Estimating weight can be done through readily captured images. Secondly, in order to deploy the model on mobile phones, we choose to research lightweight models for weight estimation, to meet the criteria of real-time estimation, portability, and low cost. Finally, we improve the accuracy of weight estimation by building three key modules.
The main contributions of this paper are as follows:
•We proposed a multi-scale keypoints generation method based on images and a novel measurement approach for measuring heart girth in cattle. These contribute to improving the accuracy of data acquisition and decision-making processes in the field of livestock farming.
•We proposed the LaWE model and deployed it on mobile phones, enabling real-time measurement of cattle weight and meeting the needs of individual cattle farmers.
•The data collection of 108 cattle from independent livestock farmers in eastern Inner Mongolia, China, including weight, height, and other measurements, provides valuable information for farmers and researchers in the field of livestock farming to make informed decisions.
The organization of this paper includes: Section 2 provides a summary of related work on non-contact measurement of livestock size and weight. Section 3 presents an overall overview of the LaWE model. Section 4 introduces the core modules and specific methods of the model. Section 5 presents the experimental analysis and results. Section 6 discusses the non-standard image case.
Related work
The research shows that the methods of measuring livestock weight by computer vision can be roughly divided into images and three dimensional reconstruction (3D). The images method uses the camera to capture RGB images of animals, and then uses the image analysis algorithm to extract features and estimate weights. On the other hand, the 3D method utilizes three-dimensional sensors or depth cameras to acquire spatial information of the animals.
Overview
To meet the needs of individual cattle farmers for real-time measurement, low cost, and low error models, we have proposed the Lightweight Cattle Weight Estimation(LaWE) model. We conducted a survey of 260 individual cattle farmers and established the goals of the model based on the survey results. According to the idea from points to lines, from lines to surfaces, and then to the whole, we have established the framework of the model.
Cattle keypoints generation module
In order to generate more accurate keypoints, we use the multi-scale keypoint coordinate generation method, including multi-scale image generation, feature extraction, scale alignment, and fusion. First, we use the Gaussian pyramid algorithm to transform the image into a multi-scale image. Then, we use MobilePoseNet to extract key feature information from multi-scale images. Finally, we perform scale alignment and fusion on the extracted key feature information to obtain high-precision.
Experiments
In this section, we perform empirical evaluations to demonstrate the effectiveness of the proposed LaWE framework. Specifically, we aim to answer the following research questions:
- •RQ1: How does the proposed LaWE model perform in mobile phones?
- •RQ2: How effective is the LaWE model in estimating results?
- •RQ3: What is the precision of the Cattle Keypoints Generation module? How does it contribute to the final estimation of the LaWE model?
- •RQ4: How does the heart girth measurement affect the accuracy
Discussion
In our study, we collected a dataset of cattle raised by individual farmers, which covers key characteristic information such as the weight, height, and so on. To validate the accuracy and practicality of the LaWE model in cattle weight estimation tasks, we conducted comparative experiments on the collected dataset and publicly available datasets. At present, although non-contact methods of measuring livestock weight.
Conclusion
In conclusion, the LaWE model constructed in this paper achieves weighing of cattle through three modules. In the Cattle Keypoints Generation module, a lightweight multi-scale keypoint generation method is proposed to obtain accurate body keypoints of cattle. In the Automated Cattle Body Measurements module, we automatically measure body measurements based on the obtained keypoints and propose a simple yet practical method for measuring the heart girth of cattle. In the Cattle Weight Prediction.







